
Research suggests that unique patterns of gene regulation have contributed to the differences in brain size and capacity that distinguish humans from other animals.
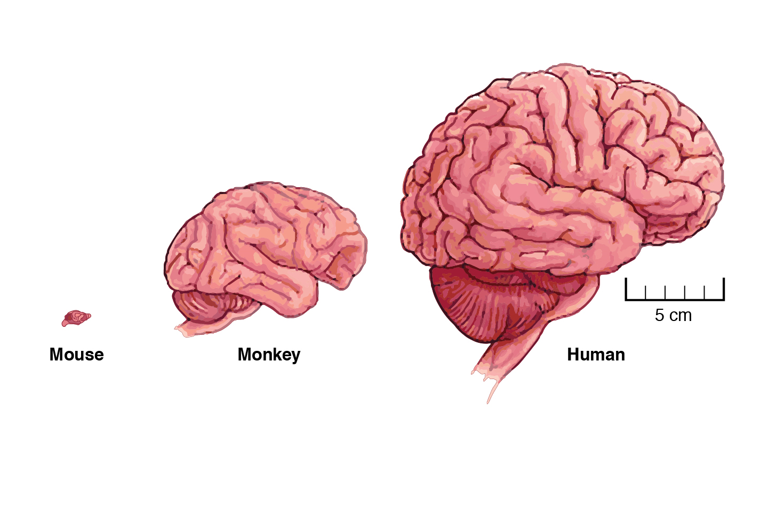
Differences in cortical size and organization underlie many of the unique capabilities that distinguish humans from other species, such as higher cognition and language. The human brain also has distinctive vulnerabilities, such as susceptibility to addiction.
How did these differences come about, when we share almost all of the genes that contribute to cortical development with other animals?
Drs. Pasko Rakic and James P. Noonan, with colleagues at Yale University, investigated an idea that evolutionary geneticists have often proposed.
Dr. Noonan explains, “It’s a long-standing hypothesis that changes in how genes are controlled, in addition to actual changes in the genes themselves, might govern evolutionary innovations such as the human cortex. The idea is that because the genes that control early brain development are very similar across species, differences in how and when these genes are expressed may distinguish the species from one another.”
Image adapted from http://image.slidesharecdn.com/031609-phitchcock-cerebralcortexlecture-110718230733-phpapp01/95/031609b-cerebral-cortex-39-728.jpg?cb=1311031041
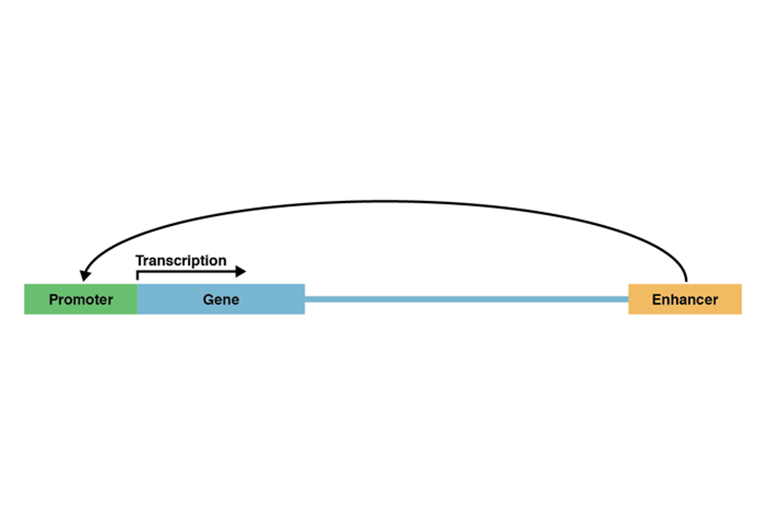
To test the hypothesis, the Yale team compared the activity levels of two regulators of gene expression in human, rhesus macaque, and mouse embryonic brain tissue. One of the regulators, called the gene promoter, is a DNA sequence located in front of a gene. When activated by the cell’s transcription machinery, it initiates the process of protein building. The other sequence, the gene enhancer, is a DNA sequence situated relatively far from a gene, in front or behind. Enhancers, too, promote gene expression when activated.
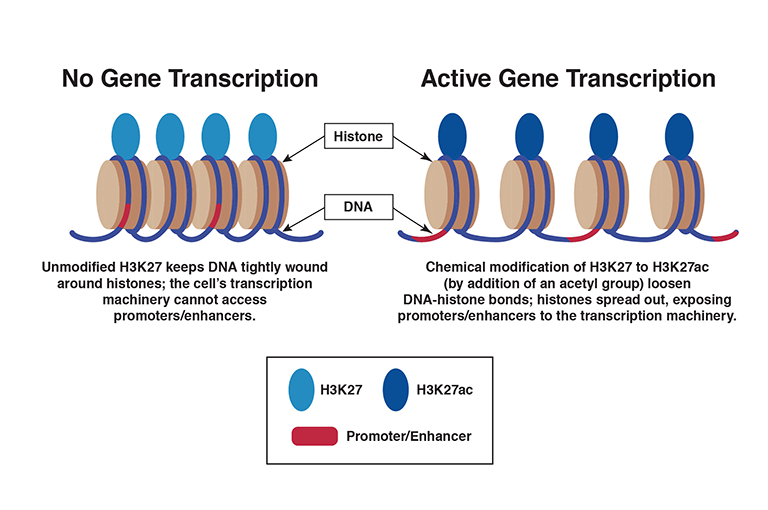
To determine the activity levels of gene promoters and enhancers, the researchers measured the levels of two proteins. The proteins, H3K27ac and H3K4me2, are modified forms of proteins (H3K27 and H3K4) that attach to histones, spool-like structures that bind with DNA.
When H3K27 and H3K4 are unmodified, DNA forms tight bonds with histones and holds them close together. Modification to H3K27ac and H3K4me2 loosens the bonds and the histones spread out, exposing DNA, including promoters and enhancers, to the transcription machinery.
Dr. Noonan says, “Simply put, the amount of [H3K27ac and H3Kme2] correlates with the activity of a promoter or enhancer.”
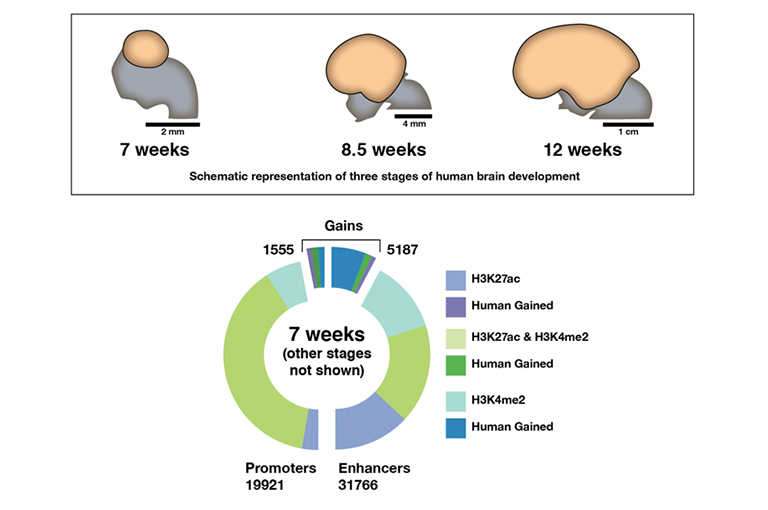
The Yale team measured H3K27a and H3Kme2 levels associated with 22,139 gene promoters and 52,317 gene enhancers in human brain tissue at 7, 8.5, and 12 weeks of embryonic development. They measured the modified proteins at the same sites in rhesus macaque monkey and mouse tissue at the analogous stages of development.
They found higher levels of one or both modified proteins adjacent to 2,885 promoters and 8,996 enhancers in humans, compared to the other species, at one or more of the three stages of development. The genes influenced by these promoters and enhancers are likely transcribed at higher rates in humans than in the other species during those developmental stages.
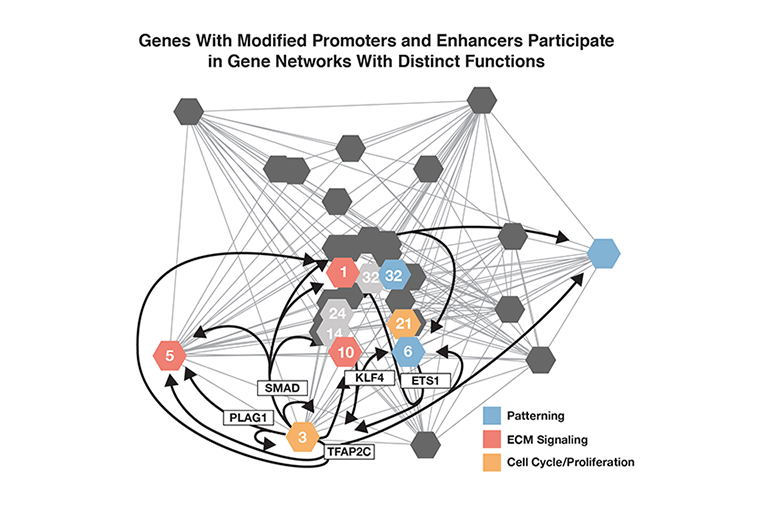
The Yale researchers identified many of the genes whose promoters and/or enhancers were associated with higher levels of H3K27ac or H3K4me2, and therefore are likely more active in humans relative to the other species. Many belong to networks with known roles in early brain development, for example:
- Cell proliferation—the birth of neurons from stem cell precursors
- Extracellular matrix building—laying in the structure that guides individual neurons to their final positions in the brain
- Patterning—the development of distinct functional brain areas
The researchers propose that these and other differences in gene regulation have helped the human brain evolve its unique size, structure, and power.
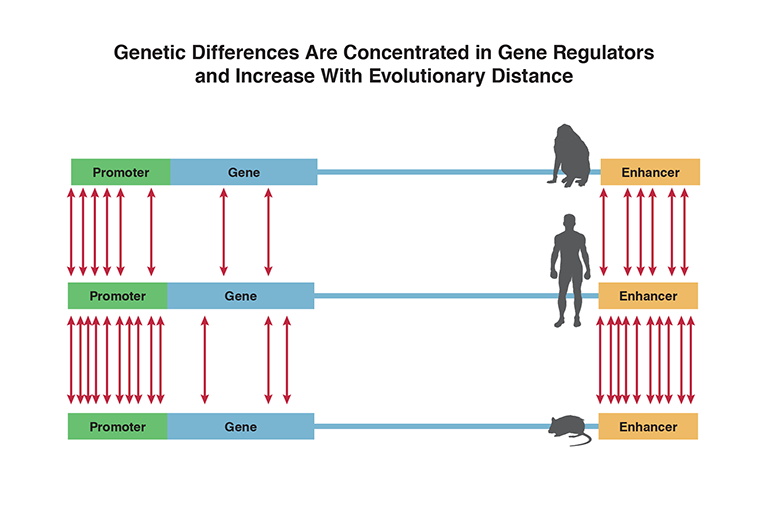
Most genetic differences between species are in gene regulators, such as promoters and enhancers, rather than in genes themselves. Differences are more numerous between species of greater evolutionary distance from each other. Dr. Rakic summarizes, “Most of the genome is conserved across species. Less than 0.5 percent of our genes are unique to humans. Most of the differences we see between species are in the regulatory DNA, including promoters and enhancers. These elements are more numerous than the genes themselves and determine the course of an organism’s development.”
Along with the increased size and capacities of our brain, our unique human patterns of gene regulation and expression may contribute to some of our weaknesses. Dr. Rakic says that our vulnerability to addiction is an example. He explains, “Something in the human brain makes us prone to addiction. This vulnerability isn’t present in other species to the same extent. Identifying genetic differences between species, including differences in regulatory DNA sequences, can inform our understanding of why we’re prone to addiction.”
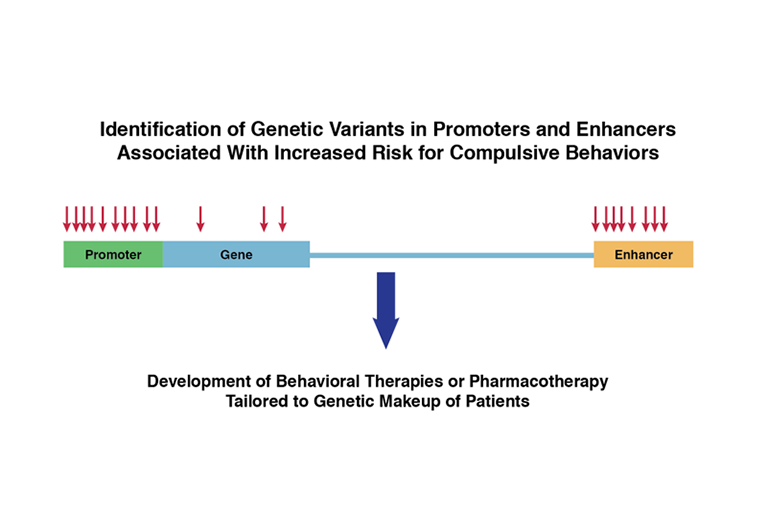
Dr. Da-Yu Wu of NIDA’s Genetics Workgroup speculates that the Yale team’s findings may, in time, have a significant impact on addiction research and treatment. He notes that substances of abuse may affect the genes driven by the human-specific promoters and enhancers associated with brain development. Therefore, these genes and their regulators could represent new therapeutic targets. Dr. Noonan finds this idea intriguing, “It’s possible, because pharmacologic agents can alter gene expression and epigenetic states in living cells.”
Dr. Wu also speculates that the researchers’ insights could guide the development of precision—or even personalized—medications for addiction treatment. Dr. Noonan elaborates, “Regulatory maps of brain development could help us interpret variations in promoters and enhancers, and perhaps other noncoding DNA regions, that are associated with addiction risk. Genetic risk factors may converge in particular regulatory pathways. If we could determine this, we might be able to identify the biological processes that are affected.”
This study was supported by NIH grants DA023999, NS014841, GM094780, and GM106628.
Source: Reilly, S.K.; Yin, J.; Ayoub, A.E.; Emera, D.; Leng, J.; Cotney, J.; Sarro, R.; Rakic, P.; Noonan, J.P. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015 Mar 6;347(6226):1155-9. Article
