Human immunodeficiency virus type-1 (HIV-1) is an infectious virus that persists in the human body even with treatment. HIV continues to replicate at low levels in the infected cells and rebounds promptly when the treatment is interrupted. People with substance use disorders (SUD) can be at high risk for contracting this virus, either through injection drug use, or risky behaviors prompted by impaired judgment. While there are medications that can help people with HIV/AIDS lead long and productive lives, there is still much scientists do not know about how to block the virus from replicating, a process called transcription. Each viral protein carries its own code—or transcript—and for the virus to thrive, these transcripts must be replicated.
In transcription, information in a strand of DNA is copied into a new molecule of messenger RNA (mRNA). This new research suggests that the addition of 5-methylcytosine (m5C) residues—a methylated form of the DNA base cytosine that regulates gene transcription—can modify the HIV-1 genetic profile. More specifically, when cytosine is mixed with the simple alcohol methanol (and is therefore methylated) the DNA maintains the same genomic sequence, but the expression of methylated genes can be altered. This kind of science is broadly called epigenetics— the study of functional changes in the regulation of gene activity and expression that are not dependent on the gene sequence. Similarly, the field of viral epitranscriptomics is the study of RNA modifications in viral transcripts that do not affect the sequence of the transcript but are functionally relevant.
Looking for opportunities to interrupt and/or better regulate the HIV transcription process, scientists at Duke University and the University of North Carolina wanted to learn more about molecular modifications of mRNA ribonucleotides—part of the basic building blocks of genetic messages. One issue has been the difficulty in quantifying these modifications. There are several techniques that can be used to map epitranscriptomic changes on RNAs but they do not provide enough information about the level of modifications at specific mapped sites. To quantify the modifications present on a single human RNA species, the investigators focused on the RNA genome of HIV-1, which is packaged into viral particles released by infected cells, and more easily purified for study than are cellular mRNAs. They then harvested material from HIV-1-infected cultures of the human CD4+ T cell line CEM and isolated the particles away from cellular debris.
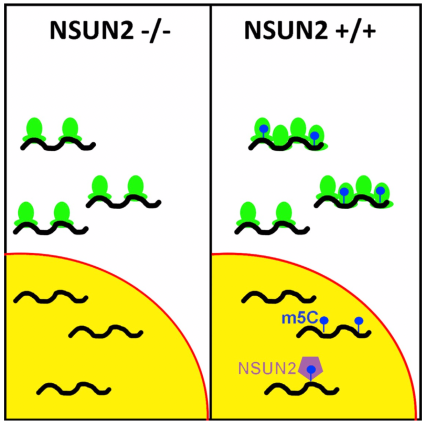
Investigators discovered that the addition of m5C in infected cells highly modifies HIV-1 mRNA, and that NSUN2 methyltransferase–a protein coding gene—adds the m5C to the nucleus. In addition, the loss of NSUN2 prevents the addition of m5C and hence inhibits decoding of HIV-1 mRNA, disrupting transcription. Overall, these findings could present an opportunity for interrupting the HIV-1 viral replication process, the first step in developing a potential cure for this disease.
Study:
- David G. Courtney, Kevin Tsai,Hal P. Bogerd, ..., Ronald Swanstrom, Christopher L. Holley, Bryan R. Cullen. Epitranscriptomic Addition of m5C to HIV-1Transcripts Regulates Viral Gene Expression. Science Direct. August 14, 2019